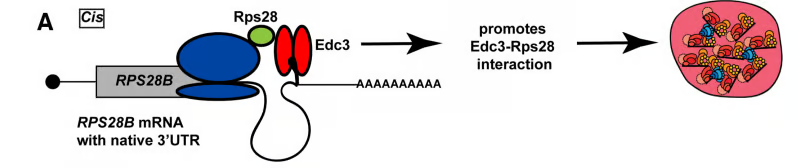
How protein-protein interactions are correctly and efficiently established is not straightforward given the complexity of the cellular milieu, off target interactions, barriers to diffusion and other factors. Spatiotemporally regulated mechanisms for facilitating the co-translational establishment of protein interactions have been described, but how widespread such mechanisms are is unclear. Recently, we described the first example in yeast of an mRNA’s 3’UTR playing a role in aiding interaction of a nascently translated protein (Rps28b) with a binding partner (Edc3), which in turn helps drive the formation of P-bodies. These findings mimic similar observations for different mRNAs and proteins by the Mayr lab in mammalian cells (see additional links below).
We are interested in determining how prevalent mRNA 3’UTR-based scaffolding of nascent protein interactions is and understanding the key mechanisms underlying this process.
Additional links:
https://www.mskcc.org/research/ski/labs/christine-mayr/overview